SPLAT
Edward Lau Lab, University of Colorado School of Medicine

SPLAT
SPLAT (Simultaneous Proteome Localization and Turnover) is a method to simultaneously quantify protein translocation and turnover rates in vivo. This repository provides a workflow to process tandem mass tag (TMT) and stable isotopic labeling by amino acids in cell culture (SILAC) mass spectrometry data generated in SPLAT experiments.
The SPLAT workflow is implemented in Snakemake and calls two in-house software packages, RIANA and PyTMT, for processing the SILAC (turnover) and TMT (localization) data, respectively. The workflow is can be run locally on a single machine.
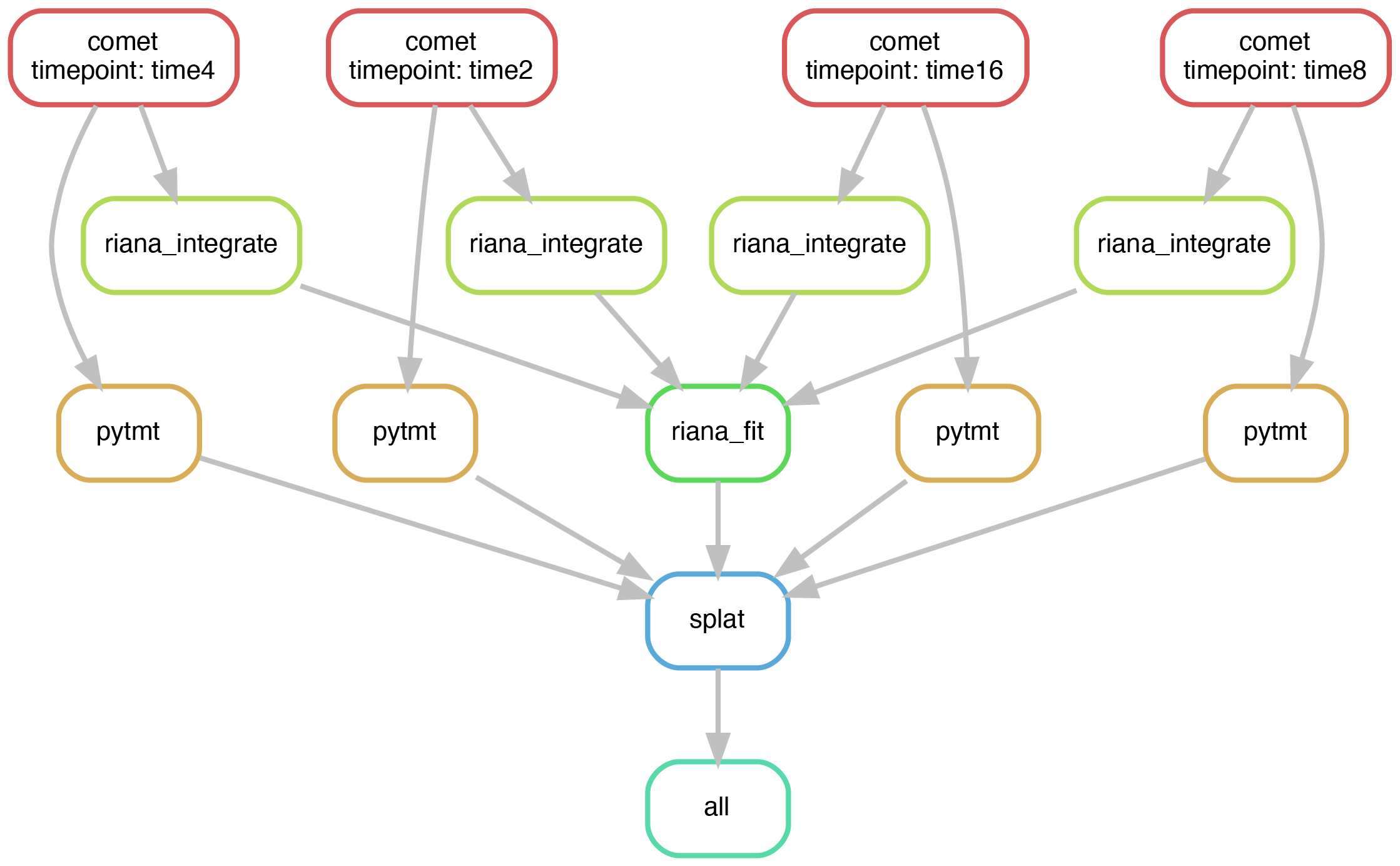
The latest version and source code of SPLAT can be found on github: https://github.com/lau-lab/splat.
See the Documentation page for instructions.
Contributors
- Edward Lau, PhD - ed-lau